NEP Model
The NEP model was initially implemented in the GPUMD software package (2022b NEP4 GPUMD). NEP training in GPUMD uses the separable natural evolution strategy (SNES), which is simple to implement as it does not rely on gradient information. However, for standard supervised learning tasks, especially deep learning, gradient-based optimization algorithms are more suitable. We implemented the NEP model (NEP4, with the network structure shown in Figure 1) in the PWMLFF 2024.5 version, which can use the gradient-based LKF or ADAM optimizers in PWMLFF for model training.
We compared the training efficiency of LKF and SNES optimization methods in various systems. The test results show that the LKF optimizer exhibits superior training accuracy and convergence speed in training the NEP model. The NEP model's network structure has only a single hidden layer, which provides very fast inference speed, and introducing the LKF optimizer significantly improves training efficiency. Users can obtain high-quality NEP models in PWMLFF at a lower training cost and use them for efficient machine learning molecular dynamics simulations, which is very friendly for users with limited resources/budgets.
We have also implemented the Lammps molecular dynamics interface for the NEP model, supporting CPU or GPU devices. Thanks to the simple network structure of NEP and the simplified feature design, the NEP model has very fast inference speed in Lammps.
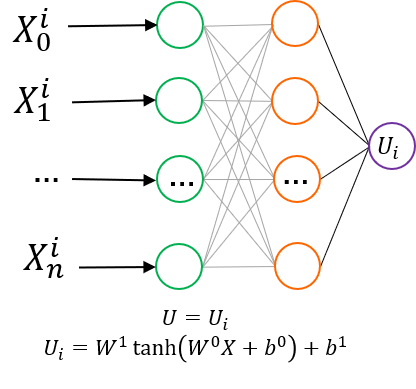
NEP network structure, where different types of elements have independent but structurally identical sub-neural networks. Additionally, unlike the NEP4 network structure in the literature, for each layer's bias, all sub-networks do not share the final layer's bias. Furthermore, we use the same cutoff for multi-body descriptors as for two-body descriptors.
NEP Command List
traintraining command, the same as for DP, NN, and Linear models. For detailed usage, refer to NEP Model Training.
PWMLFF train nep.json
- Python testing interface: We provide two commands,
inferandtest, which are used in the same way as DP inference.
For the infer command, you only need to specify the NEP potential file path. This supports nep.txt files from GPUMD, PWMLFF nep_model.ckpt potential files, as well as the nep_to_lmps.txt format file used in LAMMPS.
For example:
PWMLFF infer nep_model.ckpt atom.config pwmat/config
PWMLFF infer gpumd_nep.txt 0.lammpstrj lammps/dump Hf O
# Hf and O are the element names in the LAMMPS dump format, where Hf is element type 1 and O is element type 2 in the structure.
In the test command's test.json, the above-mentioned potential formats are also supported:
PWMLFF test nep_test.json
toneplmpscommand is used to convert thenep_model.ckptfile trained byPWMLFFinto anep_to_lmps.txtfile for use in LAMMPS simulations:
PWMLFF toneplmps nep_model.ckpt
togpumdcommand is used to convert thenep_model.ckptfile trained byPWMLFFinto anep_to_gpumd.txtfile for use inGPUMDsimulations.
The difference between nep_to_lmps.txt and GPUMD’s nep_to_gpumd.txt is that nep_to_lmps.txt additionally stores the last bias for each element type, so there are extra rows of values, where is the number of element types. If it’s a single-element system, the files will be the same.
Since the last bias is shared across all element types in GPUMD, it needs to be adjusted based on the system being simulated. The conversion logic we use is shown in the formula below:
Where is the number of element types, is the bias for element type in the potential file, and is the number of atoms of element type in the system to be simulated.
# You can enter the following command to query the input parameter details:
PWMLFF togpumd -h
# A complete example of conversion parameters is shown below, using the HfO2 system as an example. Suppose you are simulating a system with N Hf atoms and M O atoms.
# After executing the command, you will get a `nep_to_gpumd.txt` potential file, which can be used in GPUMD simulations.
# Note that this method only applies to MD simulations where the number of atoms of different types in the system does not change.
PWMLFF togpumd -m nep_model.ckpt -t Hf O -n N M
topwmlffcommand is used to convert a text-formatnep.txtpotential file into a PWMLFF Pytorch format potential file. After the command is completed, you will get a Pytorch format potential file namednep_from_gpumd.ckpt.
PWMLFF topwmlff nep.txt nep.in
NEP Model Training
Here, we use an HfO2 dataset as an example, located at [PWMLFF_nep/example/NEP].
Input File Setup
The input file for training the NEP model is similar to that for Linear\NN\DP models. You need to prepare an input control JSON file. The simplest input setup is shown below, specifying the model type as NEP, the sequence of atomic types, and the list of MOVEMENT files or pwdata format data for training. LKF optimizer will be used for training. Here, we take the pwdata format dataset as an example.
{
"model_type": "NEP",
"atom_type": [8, 72],
"max_neigh_num": 100,
"datasets_path": [
"./pwdata/init_000_50/",
"...."
]
}
If you need to use MOVEMENT format, please use the following input setup:
{
"model_type": "NEP",
"atom_type": [8, 72],
"raw_files": [
"./mvm_19_300/MOVEMENT",
"..."
]
}
It is recommended to convert the data to pwdata format before training. You can use the pwdata conversion tool or the to_pwdata conversion command provided in pwact active learning, as shown below.
# pwact command to convert MOVEMENT to pwdata format
pwact to_pwdata -i mvm_init_000_50 mvm_init_001_50 mvm_init_002_50 -s pwdata -f pwmat/movement -r -m -o 0.8
# -i list of structure files
# -f structure file format, supports pwmat/movement or vasp/outcar
# -s name of the save directory
# -r specifies saving the data in a shuffled order
# -m specifies merging the converted data, if your MOVEMENT file list has the same types and numbers of elements, you can use this parameter to save the training set as a single folder
# -o specifies the split ratio of training and test sets, default is 0.8
If your conda environment does not have pwact installed, use the following command to install it online, or refer to the pwact manual.
pip install pwact
NEP Configuration Parameters
For detailed explanations of NEP parameters, please refer to the NEP Parameters Manual.
Training
To train the NEP model, users only need to execute the following command in the directory where train.json is located. Taking [PWMLFF_nep/example/NEP] as an example:
cd /example/NEP
PWMLFF train train.json
# If you execute sbatch train.job, please modify the environment variables in the job file to your own environment variables
Output Files After Training
After training is completed, a model_record directory will be generated in the current directory, containing the following 5 files:
nep_to_lmps.txt, nep_model.ckpt, epoch_valid.dat, epoch_train.dat
nep_model.ckpt is the PWMLFF format potential file after training, and nep_to_lmps.txt is the lammps version of the potential file.
epoch_valid.dat and epoch_train.dat are loss record files during the training process.
Python Inference
The usage is exactly the same as in Python inference for DP. You just need to replace the potential file with the NEP potential file.
NEP for Lammps
We provide the Lammps interface for NEP, supporting simulations on CPU (single-core or multi-core) or GPU (single or multiple cards). You can refer to the examples/nep_lmps/hfo2_lmps_96atoms case under the lammps directory. In addition, our Lammps interface also supports NEP4 models trained by GPUMD.
Input File Setup
To use the nep_model.ckpt potential file generated after training for lammps simulations, you need to:
Step 1 Extract the potential file, simply input the following command:
PWMLFF toneplmps nep_model.ckpt
After a successful conversion, you will get a potential file nep_to_lmps.txt. If your model is successfully trained, there will be an already converted nep_to_lmps.txt file in the same directory as nep_model.ckpt, which you can directly use for lammps.
Step 2 Lammps input file setup
To use the potential file generated by PWMLFF, a lammps input file example is shown below:
pair_style pwmlff 1 nep_to_lmps.txt
pair_coeff * * 8 72
For more flexible use, we allow the order of atomic types in your potential to be different from the order of types in the lammps input structure. For this, you need to specify the atomic type corresponding to the atomic number in the simulation structure in pair_coeff. For example, if 1 in your structure is O element and 2 is Hf element, you can set pair_coeff * * 8 72.
You can also replace the 'nep-to_1mps. txt' file with the NEP4 force field file obtained from your GPUMD training.
Step 3 Start lammps simulation
If you need to use the CPU device for lammps simulation, please input the following command. Here, 64 is the number of CPU cores you need to use, please set it according to your own device.
mpirun -np 64 lmp_mpi -in in.lammps
We also provide a GPU version of the lammps interface, please input the following command. Here, 12 is the number of CPU cores you need to use. We will distribute the 12 threads evenly across the 4 GPUs you use for calculations. We recommend not using more than twice the number of CPU cores as the number of GPUs you set, as too many threads on a single GPU will reduce running speed due to resource competition.
mpirun -np 12 lmp_mpi_gpu -in in.lammps
About Lammps Environment Variables
To run lammps, you need to import the following environment variables, including your PWMLFF path and lammps path.
# Import cuda
module load cuda/11.8 intel/2020
# Import your conda environment variables
source /the/path/anaconda3/etc/profile.d/conda.sh
conda activate torch2_feat
# Import your PWMLFF environment variables
export PATH=/the/path/codespace/PWMLFF_nep/src/bin:$PATH
export PYTHONPATH=/the/path/codespace/PWMLFF_nep/src/:$PYTHONPATH
# Import your lammps environment variables
export PATH=/the/path/codespace/lammps_nep/src:$PATH
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:$(python3 -c "import torch
; print(torch.__path__[0])")/lib:$(dirname $(dirname $(which python3)))/lib:$(dirname $(dirname $(which PWMLFF)))/op/build/lib
NEP Model Test Results
LKF Optimizer is More Suitable for NEP Models
We have tested multiple systems, using 80% of the dataset as the training set and 20% as the validation set. We trained the NEP model on a public HfO2 training set (including 2200 structures of 𝑃21/c, Pbca, 𝑃ca21, and 𝑃42/nmc phases) with LKF and evolutionary algorithms (SNES, GPUMD). Their error reduction on the validation set is shown in Figure 2. With the increase of training epochs, the NEP model based on LKF converges to a lower error level (lower error indicates higher accuracy) faster than SNES. Similar results are observed in the aluminum system (including 3984 structures) (Figure 3). Moreover, similar results are found in the LiGePS system and quinary alloy system. For more detailed data, please refer to the uploaded training and test data.
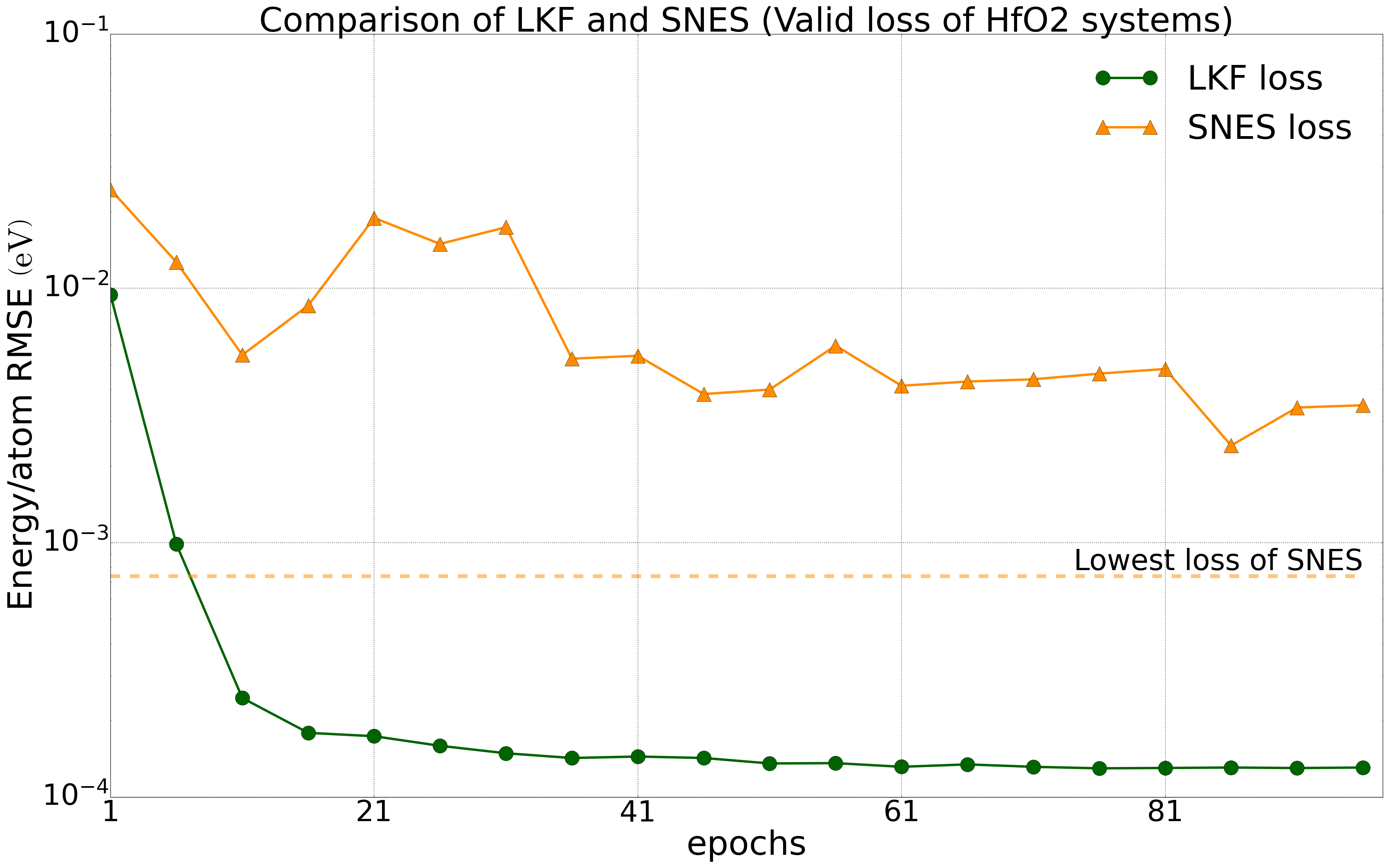
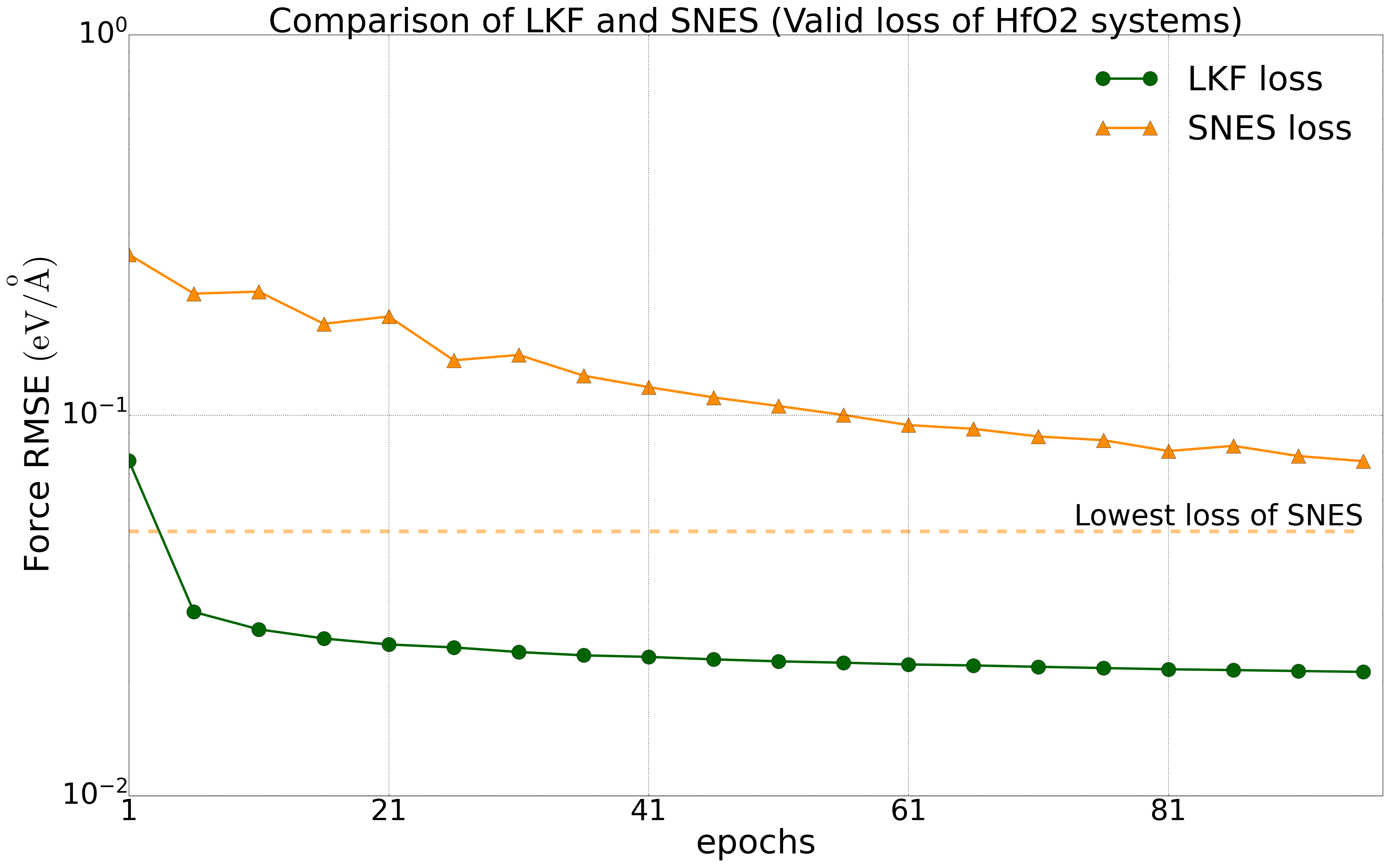
Convergence of energy (left) and force (right) for the NEP model under LKF and SNES optimizers in the HfO2 system (2200 structures). The dashed line indicates the lowest loss level that can be achieved by SNES algorithm training.
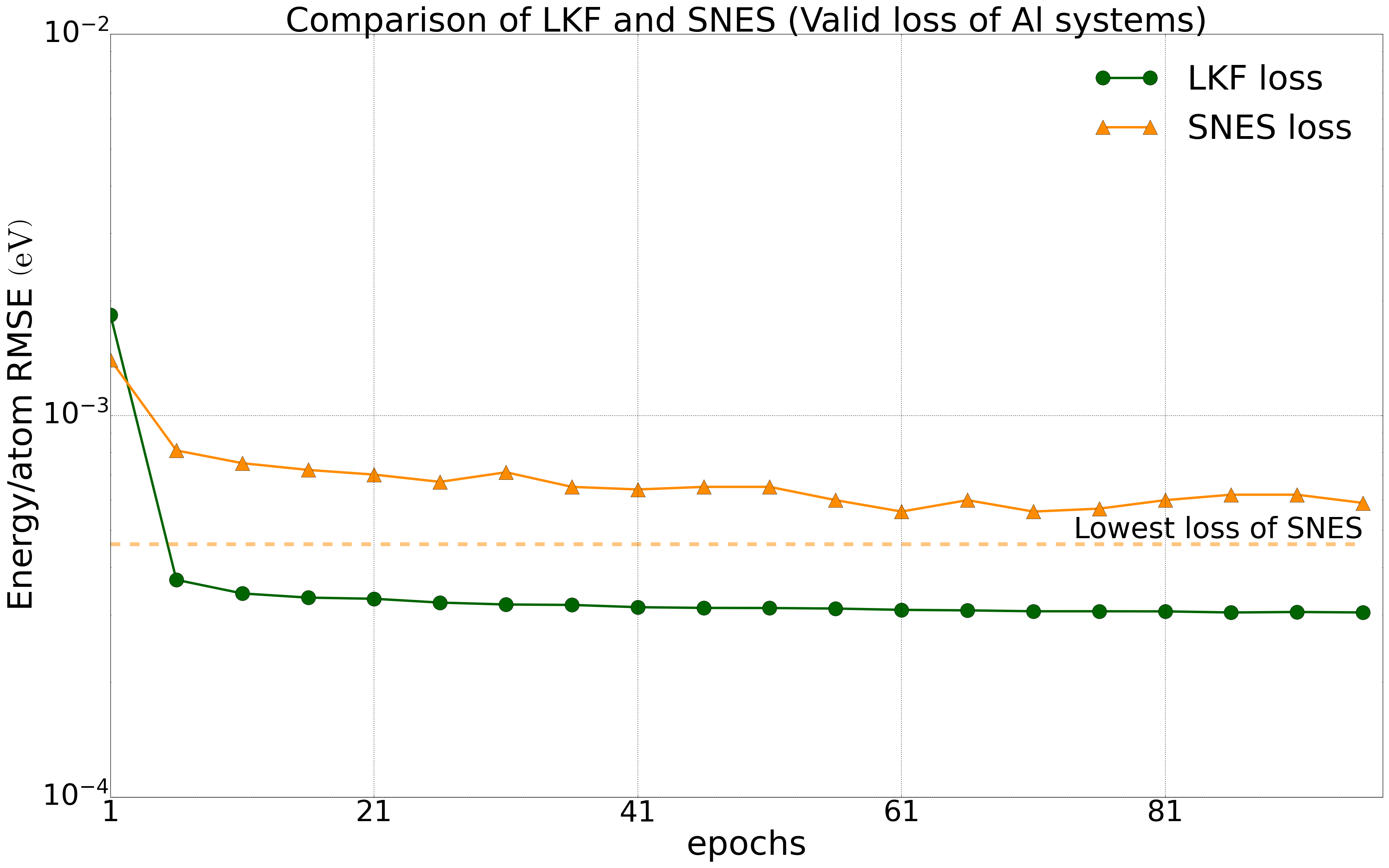
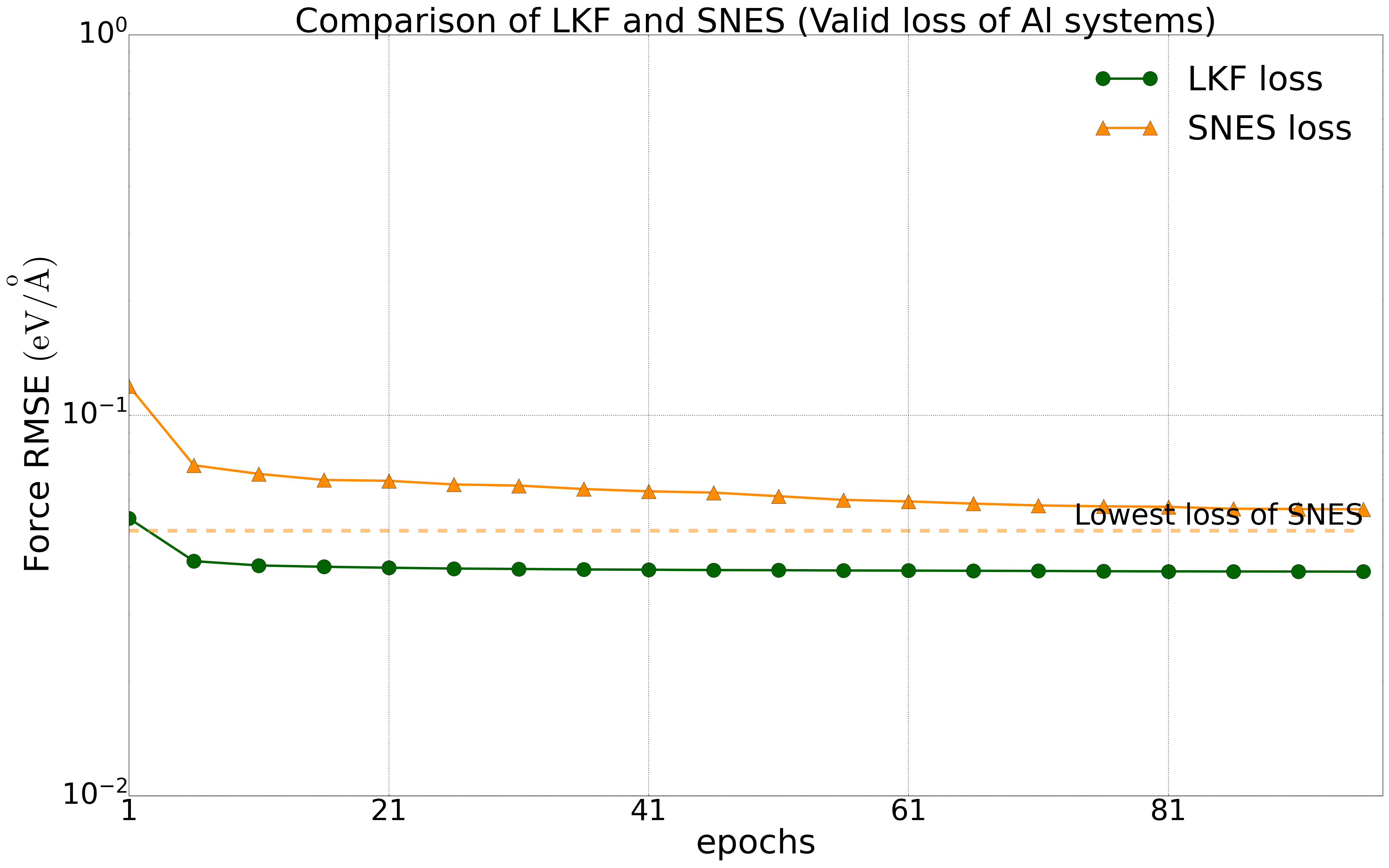
Convergence of energy (left) and force (right) for the NEP model under LKF and SNES optimizers in the Al system (3984 structures). The dashed line indicates the lowest loss level that can be achieved by SNES algorithm training.
Accuracy Comparison Between NEP and Deep Potential Models in PWMLFF
The deep potential (DP) model is a widely used neural network model, and PWMLFF implements a Pytorch version of the DP model, which can also use the LKF optimizer. We compared the training of NEP and DP (PWMLFF) models using the LKF optimizer in multiple systems, as shown in Figure 4. In Al, HfO2, LiGePS (including 10,000 structures), and [Ru, Rh, Ir, Pd, Ni] quinary alloy systems (including 9486 structures), the NEP model in PWMLFF converges faster and achieves higher accuracy than the DP model. Specifically, for the quinary alloy, we used type embedding DP to reduce the impact of the number of elements on training speed (in previous tests, we found that for situations with more than five elements, introducing type embedding in PWMLFF's DP training can achieve higher accuracy than regular DP).
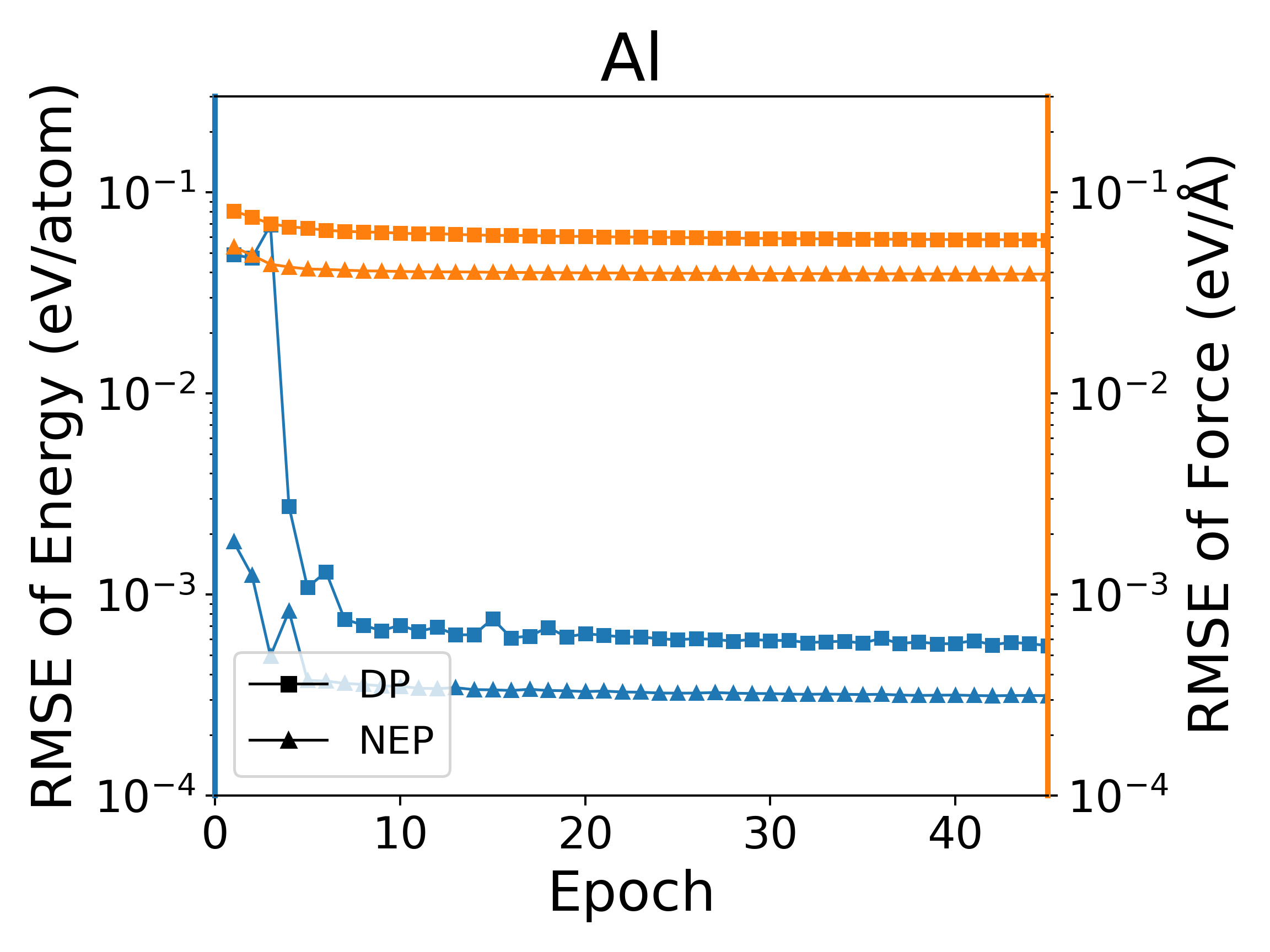
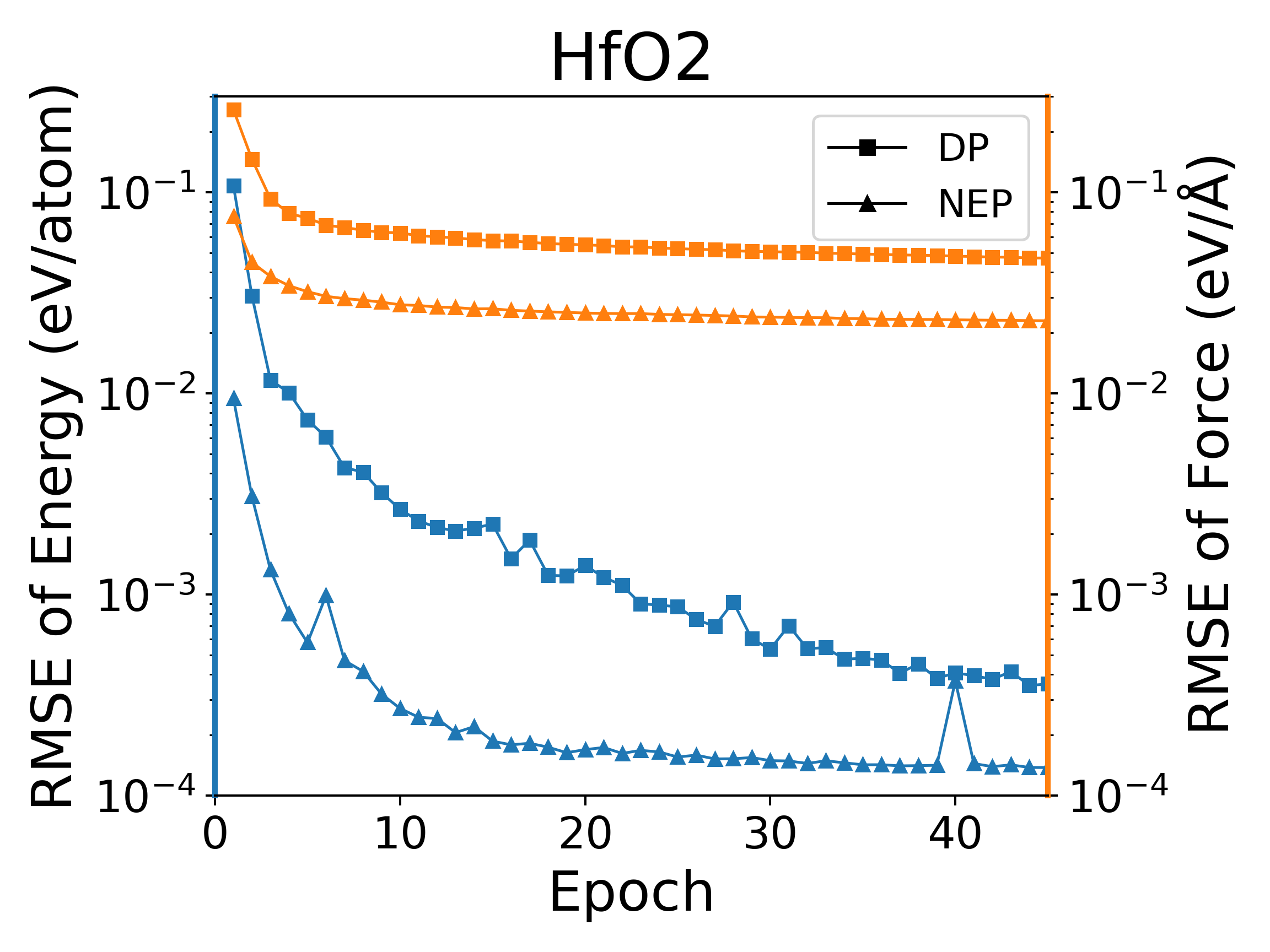
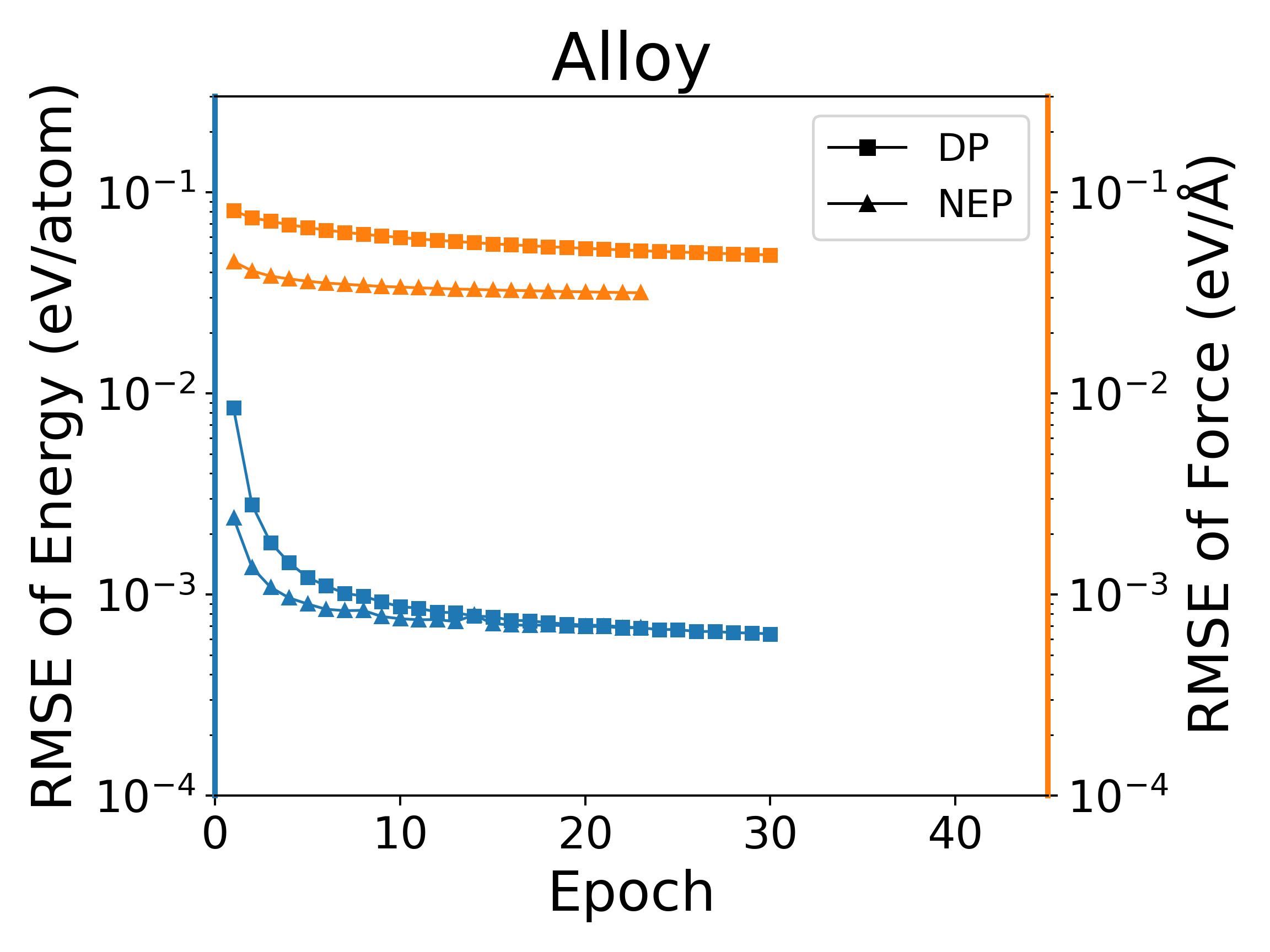
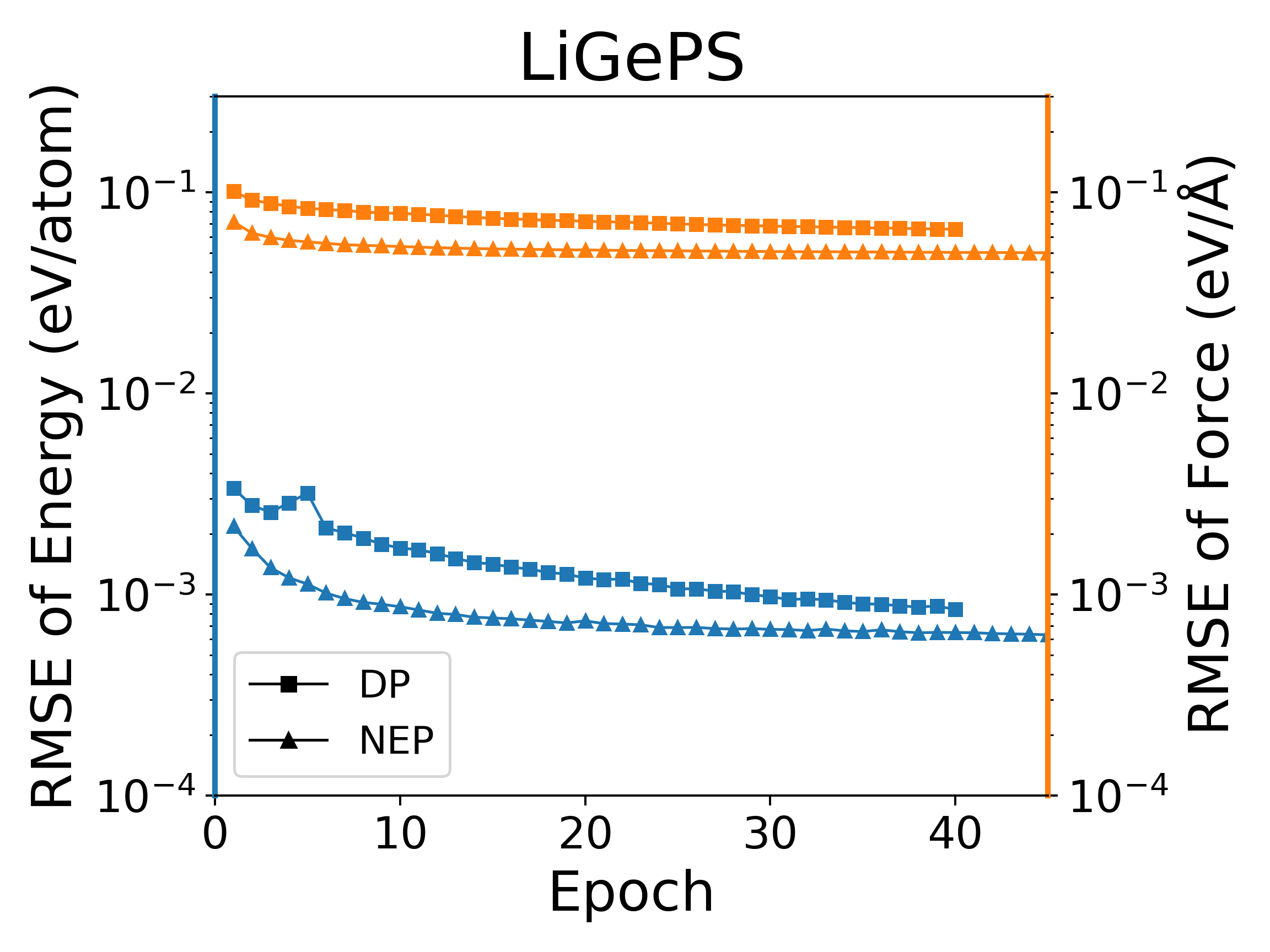
NEP and DP model training error convergence under LKF optimizer
Test Data
Test data and models have been uploaded. You can download them from our Baidu Cloud Disk https://pan.baidu.com/s/1beFMBU1IehmNEpIQ9B8ybg?pwd=pwmt, or from our open-source dataset repository.
Lammps Interface Test Results
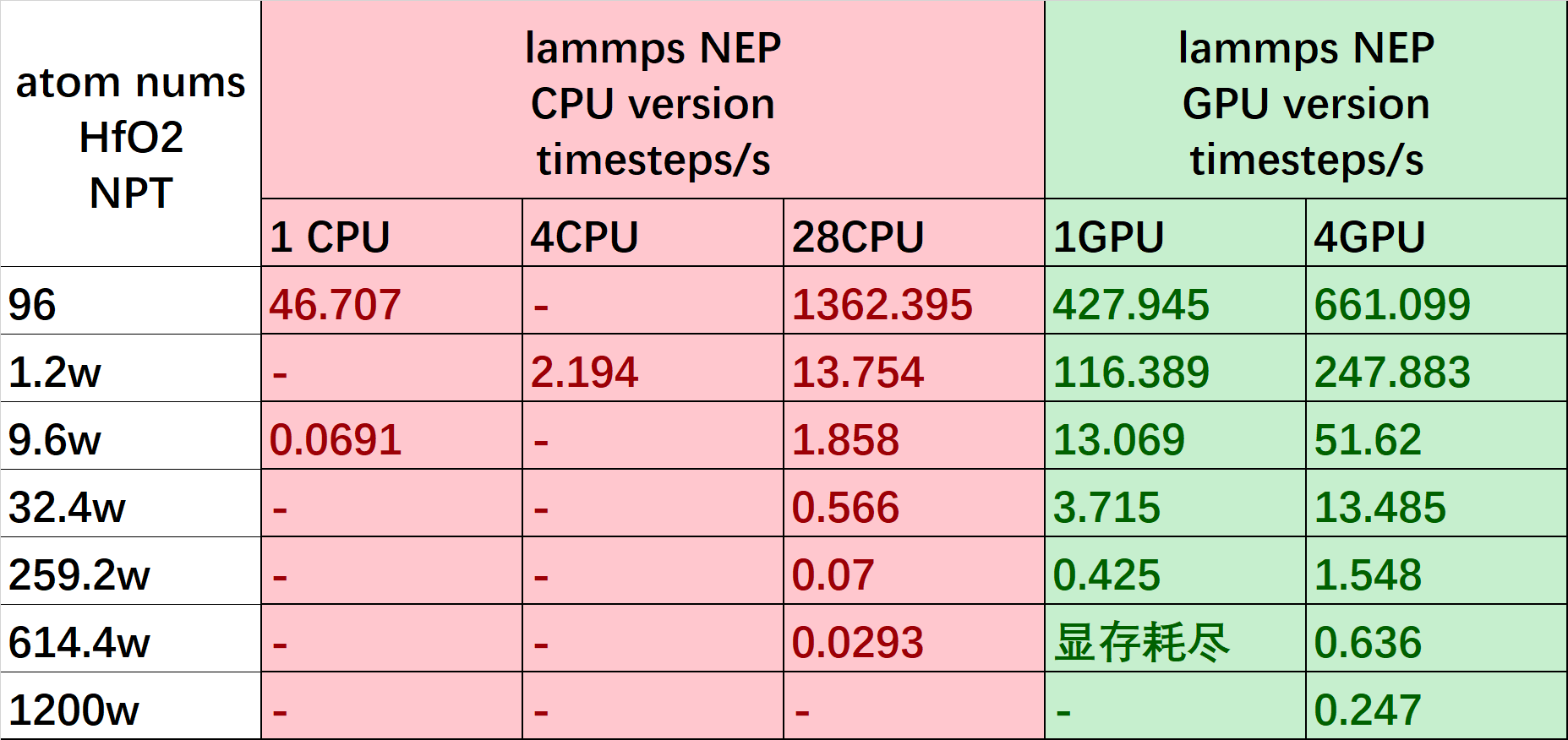
The following figure shows the simulation speed of the NEP model's lammps CPU and GPU interface for NPT ensemble MD simulations on a 3090*4 machine. For the CPU interface, the speed is proportional to the atomic scale and the number of CPU cores; for the GPU interface, the speed is proportional to the atomic scale and the number of GPUs.
Based on the test results, we recommend using the CPU interface if your system size is below the order of . Additionally, when using the GPU interface, we recommend using the same number of CPU cores as the number of GPU cards you set.